Getting started¶
The G-Node REST-API uses the odML data model for metadata and the neo data model for electrophysiology data. Therefore those models are also used in this client library. In order to get a better understanding about both concepts it is recommended to familiarize yourself with the neo documentation and the publication about odML: A bottom-up approach to data annotation in neurophysiology.
The best way to follow this getting started guide ist to start python in interactive mode or ipython and try out the following code examples.
Before you can use the G-Node client, you have import the gnodeclient package and connect to an instance of a G-Node server. If you don’t have a local installation you may use credentials from the example below. Since we need the neo package later on, we also include this.
1 2 3 4 | import neo
from gnodeclient import session, Model
s = session.create(location="http://test.g-node.org", username="demo", password="demo")
|
Once you have a session object and the session object is open, you can retrieve some data from the G-Node server.
1 | blocks = s.select(Model.BLOCK, {"max_results": 10})
|
The variable blocks now contains the first ten neo.Block objects that are owned by or shared with you. The method Session.select() is used to query the G-Node REST API for objects of a certain type. The type is determined by passing the model name of the object as first parameter to the Session.select() method. Furthermore select accepts filters, which can be used filter the results by certain criteria.
The class Model provides a set of constants representing each model name. The following table contains all supported types with their respective names and constants.
| Type | Name | Constant |
|---|---|---|
| odml.Section | “section” | Model.SECTION |
| odml.Property | “property” | Model.PROPERTY |
| odml.Value | “value” | Model.VALUE |
| neo.Block | “block” | Model.BLOCK |
| neo.Segment | “segment” | Model.SEGMENT |
| neo.EventArray | “eventarray” | Model.EVENTARRAY |
| neo.Event | “event” | Model.EVENT |
| neo.EpochArray | “epocharray” | Model.EPOCHARRAY |
| neo.Epoch | “epoch” | Model.EPOCH |
| neo.RecordingChannelGroup | “recordingchannelgroup” | Model.RECORDINGCHANNELGROUP |
| neo.RecordingChannel | “recordingchannel” | Model.RECORDINGCHANNEL |
| neo.Unit | “unit” | Model.UNIT |
| neo.SpikeTrain | “spiketrain” | Model.SPIKETRAIN |
| neo.Spike | “spike” | Model.SPIKE |
| neo.AnalogSignalArray | “analogsignalarray” | Model.ANALOGSIGNALARRAY |
| neo.AnalogSignal | “analogsignal” | Model.ANALOGSIGNAL |
| neo.IrregularlySampledSignal | “irregularlysampledsignal” | Model.IRREGULARLYSAMPLEDSIGNAL |
Since the G-Node client returns only slightly extended versions of the native neo and odML objects, working with them is quite simple. Lets examine the block a bit closer:
1 2 3 4 | block = blocks[0]
print block.name
print block.description
print block.location
|
As normal neo.Block object the block returned by the session has a name and a description. But in addition it has also a property called location. This is one of the minor extensions that are introduced by the client. The location is an identifier that allows the client library to identify the corresponding remote entity of the object.
Additionally to normal properties Block objects have relationships to other objects like Section, Segment and RecordingChannelGroup. For objects returned by methods of the Session class, properties representing those relationships are initialized with lazy-loading proxies. This means, that related objects are only downloaded or fetched from the cache, when the respective properties of the object are accessed for the first time. The following piece of code illustrates this behaviour.
1 2 3 | print type(block.segments)
print len(block.segments)
print len(block.recordingchannelgroups)
|
The output of line one will show, that Block.segments is a proxy object. As soon as data from the proxy is requested (line 2 and 3) the data will be fetched from the server or the cache.
The Session.select() method is used to get data by type and provides the possibility to reduce the results by filter. A second method for getting data is Session.get(). This method takes a single object identifier, the location, as first argument.
1 | block = s.get(block.location, refresh=True)
|
The parameter refresh controls whether or not the client should check for updates on the server if the object was found in the cache.
The next code example demonstrates how to upload data to the G-Node server via REST API. The Session.set() method can be used to upload a new neo or odML object to the server or to update the state of an existing object. The code in the listing first creates a new neo.Segment and adds this segment to the to the segment list of an existing block before it is uploaded to the server using Session.set().
1 2 3 4 5 6 | segment = neo.Segment("cool segment")
segment.block = block
segment = s.set(segment)
block = s.get(block.location, refresh=True)
|
The above example reveals some design principles of the G-Node API and the client library:
- Associations between objects can only be changed on the child side of the one-to-many relationship.
- All functions of the client interface have pure input parameters. Further more previously returned objects are never changed by subsequent function calls. In this example the content of block.segments changes on the server when the segment was saved using Session.set(). But since the local :py:attr`block` object was not changed, it block has to be updated (see line 6).
Data on the server can be accessed by type (e.g. time segment, analog signal) with filters using model attributes. For instance, according to the Neo model, analog signals have the sampling rate as an attribute. The following query requests analog signals with a certain sampling rate:
1 | signals = g.select(Model.ANALOGSIGNAL, {"sampling_rate": 500, "max_results": 5})
|
The “select” function of the client library accepts, as a second parameter, filters in a Python dict object.
Structured data can be accessed by spatial (e.g. recording channel), temporal (segment), or source (unit) criteria. The following request finds a certain recording channel and fetches all data coming from it:
1 2 | selection = g.select(Model.RECORDINGCHANNEL, {"index": 8})
channel_with_data = g.get(selection[0].location, recursive=True)
|
Here the “select” function is used to query recording channel objects having “index” attribute set to 8. Every object, fetched from the server, has a “location” attribute which allows the library to determine the corresponding remote entity of the object. Then the “get” function allows to request the first channel from the previous selection with all related data recursively (analog signals, spike trains).
Another request finds a certain unit (in this example, a neuron given number 3) and fetches all spike trains detected from it:
1 2 | selection = g.select(Model.UNIT, {"name__icontains": "3"})
unit_with_spike_data = g.get(selection[0].location, recursive=True)
|
For the organization of metadata, the client library provides an interface to the python-odml library, so that odML objects can be natively manipulated and stored to the central storage. Additionally, odML provides terminologies with predefined metadata items. Those terminologies can be loaded directly from the odML repository:
1 2 | odml_repository = "http://portal.g-node.org/odml/terminologies/v1.0/terminologies.xml"
terminologies = odml.terminology.terminologies.load(odml_repository)
|
Terminologies can be used as templates to describe certain parts of the experimental protocol. Among basic terminologies are templates for experiment, dataset, electrode, hardware configuration, cell etc. These terminologies can be accessed as a python “list” or “dict” as python-odml objects, and can be cloned to be used to annotate the current dataset:
1 | experiment = terminologies.find("Experiment").clone()
|
To describe the experiment, appropriate values are assigned to the properties:
1 2 3 4 5 6 | experiment.name = "LFP and Spike Data in Saccade and Fixation Tasks"
experiment.properties["ProjectName"].value = "Scale-invariance of receptive field properties ..."
experiment.properties["Description"].value = "description of the project"
experiment.properties["Type"].value = "electrophysiology"
experiment.properties["Subtype"].value = "extracellular"
experiment.properties["ProjectID"].value = "PMC1913534"
|
Of course, additional properties can be introduced as needed. For example, stimulus parameters can be documented using custom odML section with custom properties:
1 2 3 4 5 6 7 | stimulus = odml.Section(name="Stimulus", type="stimulus")
stimulus.append(odml.Property("BackgroundLuminance", "25", unit="cd/m2"))
stimulus.append(odml.Property("StimulusType", "SquareGrating", stimulus))
stimulus.append(odml.Property("Sizes", ["1.2", "2.4", "4.8", "9.6"], unit="deg"))
stimulus.append(odml.Property("Orientations", ["0", "45", "90", "135"], unit="deg"))
stimulus.append(odml.Property("SpatialFrequencies", ["0.4", "0.8", "1.6", "3.2"], unit="1/deg"))
stimulus.append(odml.Property("NumberOfStimulusConditions", "128"))
|
Note that instead of creating metadata objects in Python, odML metadata structures can be read from file using the standard odML library. The odML format allows nested sections to capture the logical strucuture of the experiment. For example, a stimulus can be defined as part of an experiment:
1 | experiment.append(stimulus)
|
This tree-like structure can be saved with the client library:
1 | experiment = tools.upload_odml_tree(g, experiment)
|
The client library allows searching for metadata of a particular type, using different filters that can be applied for object attributes:
1 | sections = g.select(Model.SECTION, {"name__icontains": "LFP and Spike Data"})
|
For complex experiments, the entire tree of metadata subsections can be very large. Therefore, the “select” function does not return the whole tree, instead it returns only the top level section objects with lazy-loaded relationship attributes, which will fetch related objects at the moment when they are first accessed. If the user wants to download the entire tree, it can be fetched with the “get” function with “recursive” parameter:
1 | experiment = g.get(sections[0].location, recursive=True)
|
If another, similar experiment is performed, the metadata tree can simply be cloned and only the metadata that have changed updated. This is highly convenient and saves the time of re-entering parameters that stay the same across a series of experiments.
To meaningfully annotate data by metadata, the gnodepylib allows to connect datasets with the metadata:
1 2 | block.section = experiment
block = g.set(block) # updates relationship on the server
|
Note an association between objects can only be set on one-side of the one-to-many relationship. In this case a section can have many blocks, thus the block has to be changed to establish connection.
Additionally, the client library allows to connect data and metadata with so-called annotations, to indicate certain specific attributes for any of the Neo-type objects. A typical use case for this function is to specify which stimulus was applied in each trial of the experiment. This connection is done using the “metadata” attribute that uses existing metadata properties and values to “tag” a number of data-type objects:
1 2 3 4 5 6 7 | stimulus = experiment.sections["Stimulus"]
orientation = stimulus.properties["Orientations"].values[3]
size = stimulus.properties["Sizes"].values[1]
sf = stimulus.properties["SpatialFrequencies"].values[2]
segment.metadata = [orientation, size, sf]
segment = g.set(segment) # sends updates to the server
|
Proper annotation brings more consistency in data and metadata, and allows to select data by metadata in various ways. For example, for data analysis it is often necessary to select all data recorded under the same experimental conditions. The following example selects all LFP data across all trials with a certain stimulus properties:
1 2 3 4 5 6 7 8 9 | stimulus = g.select(Model.SECTION, {"odml_type__icontains": "stimulus"})[0]
filters = {}
filters["name__icontains"] = "4"
filters["^1metadata"] = stimulus.properties["Orientations"].values[0].location
filters["^2metadata"] = stimulus.properties["Sizes"].values[0].location
segment = g.select(Model.SEGMENT, filters)[0]
signals = segment.analogsignals
|
In this example we select a section describing stimulus, and use certain values of its properties to build a required filter. This filter, containing certain stimulus orientation and size can be used to query time segments where this combination was used. This type of query makes it straightforward to, for instance, compute averages across trials for a certain stimulus configuration,
1 2 | import numpy as np
signalaverage = np.mean(signals, axis=0)
|
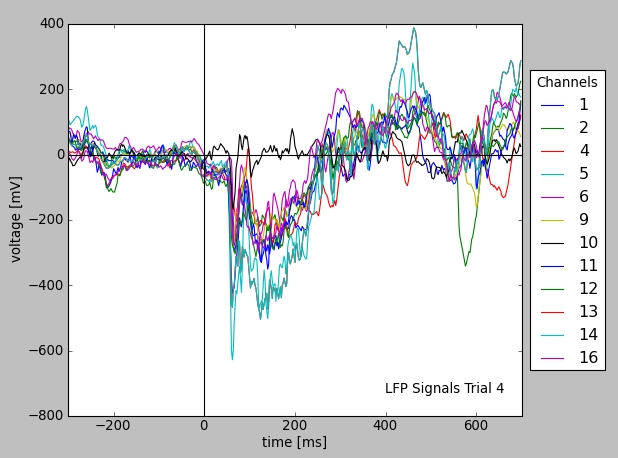
or to plot the actual LFP traces for visualization:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 | from matplotlib import pylab as pl
s1 = signals[0] # one of the signals
fig = pl.figure()
lfp = pl.subplot(111)
lfp.text(.85, .05, s1.segment.name, horizontalalignment="center",
transform=lfp.transAxes) # caption from time segment name
for s in signals:
lfp.plot(s.times, s, label=s.recordingchannel.index)
pl.xlim([s1.t_start, s1.t_stop]) # set X axis range
pl.xlabel("time [%s]" % s1.times.units.dimensionality.string) # set X units
pl.ylabel("voltage [%s]" % s1.units.dimensionality.string) # set Y units
# [...] # commands for axes and legend omitted
pl.show()
|
The figure below illustrates the resulting plot. Note that the availability of metadata with the data immediately enables meaningful labeling of the axes without having to collect further information from files or hand-written documentation.